Table 3 Characteristics of sequences implicated in lipid binding. The isolelectric point for the isolated peptide was calculated and the percent alpha-helix determined from the relevant crystal structure. The symbols for electrically positive residues are underlined (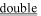 ); those corresponding to electrically negative residues are underlined (
); those corresponding to electrically negative residues are underlined ( ). The characters under the amino acid sequence refer to the secondary structure; H = helix, T = hydrogen-bonded turn, S = bend, E = extended beta-strand, and B = residue in isolated beta-bridge. Residues 401–406 (KKKKSK) are not present in talin crystal structures. Helical residues are underlined (
). The characters under the amino acid sequence refer to the secondary structure; H = helix, T = hydrogen-bonded turn, S = bend, E = extended beta-strand, and B = residue in isolated beta-bridge. Residues 401–406 (KKKKSK) are not present in talin crystal structures. Helical residues are underlined (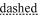 ).
).
Protein | Residues | Sequence | Number Residues | Isoelectric Point | Helix Content | Sequence Site in Protein |
|---|---|---|---|---|---|---|
α-actinin | 281–300 |
| 20 | 4.49 | 15/20 (75%) | Helices 1–2 |
720–739 |
| 20 | 4.66 | 16/20 (80%) | Carboxyl-terminal portion of Helix 16 | |
Arp2 | 185–202 |
| 18 | 10.0 | 13/18 (72%) | Helix 1 of Actin-like Subdomain 4 |
CapZβ-1 | 134–151 |
| 18 | 9.62 | 0/18 (0%) | Contains portion of β strand 6 |
215–232 |
| 18 | 4.49 | 18/18 (100%) | Helix 5 | |
Talin | 385–406 |
| 22 | 8.61 | 9/22 (41%) | Helix 5 of Subdomain F3 of Talin-H |
Vinculin | 935–978 |
| 44 | 9.73 | 31/44 (70%) | Domain 5, Helices 2–3 + amino-terminal portion of Helix 4 |
| ||||||
1020–1040 |
| 21 | 4.47 | 20/21 (95%) | Domain 5, Helix 5 | |
1052–1066 |
| 15 | Hairpin [122] |

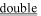 ); those corresponding to electrically negative residues are underlined (
); those corresponding to electrically negative residues are underlined ( ). The characters under the amino acid sequence refer to the secondary structure; H = helix, T = hydrogen-bonded turn, S = bend, E = extended beta-strand, and B = residue in isolated beta-bridge. Residues 401–406 (KKKKSK) are not present in talin crystal structures. Helical residues are underlined (
). The characters under the amino acid sequence refer to the secondary structure; H = helix, T = hydrogen-bonded turn, S = bend, E = extended beta-strand, and B = residue in isolated beta-bridge. Residues 401–406 (KKKKSK) are not present in talin crystal structures. Helical residues are underlined (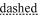 ).
).








